| Research Article | ||
J. Microbiol. Infect. Dis., (2024), Vol. 14(2): 80–87 Research Article Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, NigeriaLawrence Shola Owolabi1, Cajethan Onyebuchi Ezeamagu1*, Godswill Nduka Anyasor21Department of Microbiology, Babcock University, Ilishan, Nigeria 2Department of Biochemistry, Benjamin S. Carson (Snr.) School of Medicine, Babcock University, Ilishan-Remo, Nigeria *Corresponding Author: Cajethan Onyebuchi Ezeamagu. Department of Microbiology, Babcock University, Ilishan, Nigeria. Email: onyezecajeth [at] yahoo.com Submitted: 06/04/2024 Accepted: 20/06/2024, Published: 30/06/2024 © 2024 Journal of Microbiology and Infectious Diseases
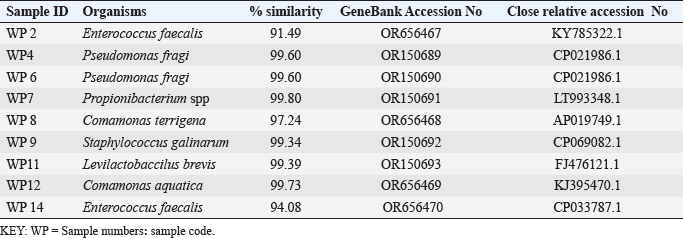
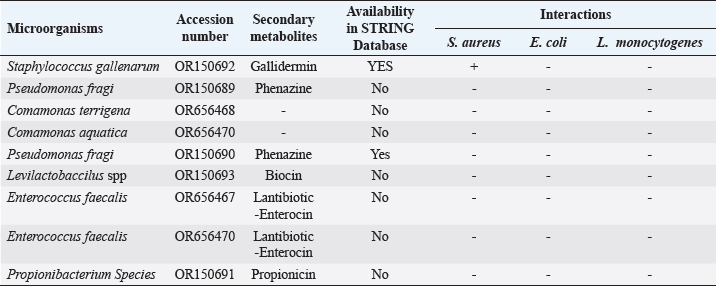
ABSTRACTBackground: Antibiotic resistance is a global problem with high morbidity and mortality in healthcare institutions that requires an antimicrobial alternative and little is known of antibacterial agents derived from insect sources. Aim: The aim of the study was to examine the antibacterial properties of secondary metabolites from insect-associated bacteria. Methods: Thirty-three wasps were obtained from abandoned structures in a local Community. Bacteria were isolated and identified by the Sanger sequencing method. The metabolites were extracted and antibacterial potential was tested against different pathogens. The metabolite concentration was analyzed via high-performance liquid chromatography (HPLC). The metabolites and their clinical potential were explored through STRING analysis. Results: Fifteen bacteria were isolated and 9 exhibited antibacterial properties consisting of Enterococcus faecalis (2). Pseudomonas fragi (2), Propionibacterium spp (1), Comamonas terrigena (2), Staphylococcus galinarum (1), Levi lactobacillus brevis (1). Antibiogram revealed that the secondary metabolite had inhibitory effects against all pathogens. The HPLC results indicated that the control and metabolites were eluted at 1.288 and 1.259 minutes, respectively, with concentrations ranging from 16.68 to 4761.82 mg/l. STRING analysis showed that the metabolites were gallidermin, enterocin, propionicin, and phenazine. Conclusion: The study concluded that the metabolites show activity against test strains and therefore a source of novel antimicrobials for addressing global drug-resistant pathogens. Keywords: Antibacterial Properties, HPLC, In-silico analysis, Secondary metabolites, Wasp-associated bacteria. IntroductionAntibacterial drug resistance has emerged as a grave global health concern, contributing significantly to morbidity and mortality in healthcare institutions worldwide (Oliver et al., 2011). Each year, antibacterial drug resistance is responsible for around 5 million fatalities, causing extended hospital stays, treatment inefficacies, and an increasing reliance on expensive second and third-line antibacterial drugs, further complicating the treatment landscape (Laxminarayan, 2022). In the United States, alarmingly high drug resistance rates have been observed in bacteria with antibiotics such as methicillin, levofloxacin, ciprofloxacin, and clindamycin (Murray et al., 2022). In Europe, multidrug-resistant bacteria are responsible for over 400,000 infections and 25,000 deaths each year (Nwobodo et al., 2022). Likewise in Sub-Saharan Africa and South Asia, the death rates per 100,000 population directly attributed to antibacterial resistance at 24%–22%, respectively. These regions also have high associated death rates (99 and 77 per 100,000) (Nwobodo et al., 2022). The prevailing antibiotic resistance made pharmaceutical companies across the globe introduce different strategies in drug development to combat the drug resistance menace. However, the antibiotic pipeline continues to dwindle, and resistance emerges to successive generations of antibiotics, including tetracycline, macrolides, aminoglycosides, ampicillin, and chloramphenicol (Imran et al., 2018). Even sulphonamides, introduced after the development of resistance to penicillin, faced resistance issues (Laxminarayan, 2022). The introduction of fluoroquinolones as powerful, broad-spectrum antibiotics was a significant milestone in the fight against resistant strains. Nevertheless, resistance to fluoroquinolones emerged, contributing to over 70% of antibiotic resistance-related deaths across all pathogens. Subsequently, the antibiotic colistin was introduced, but resistance developed rapidly. Other alternatives, including phage therapy, nano-treatment, fungi-based sources, and plant-based therapies, were explored but also faced resistance challenges (Laxminarayan, 2022). Innovative approaches are essential to address this pressing public health issue and develop alternative treatments for bacterial infections. Given the failures of antibiotic generations and alternative therapies, there is a growing need to explore novel methods for treating microbial infections. Antibacterial agents sourced from insects remain limited. Hence, exploring antibiotic properties of secondary metabolites produced from wasp-associated bacteria holds promise. Materials and MethodsResearch designThe research design was experimental involving laboratory testing and analysis. The study was conducted at the Department of Microbiology Postgraduate Laboratory, School of Science and Technology, Babcock University, Ilisan-Remo, Ogun State. Collection of samplesThirty-three matured wasps, specifically Sceliphron caementarium (black and yellow wasp), were collected from abandoned buildings in Ishara, Remo, Ogun State, between March and April 2022. The wasp nests from walls and ceilings were dislodged with a broom, followed by capturing the wasps using an aerial net. With the aid of protective hand gloves, the wasps were gently removed from the net, transferred to a sealed plastic container, and transported to the Microbiology laboratory at Babcock University. Microbial isolationMicrobial isolation was conducted using Streptomyces agar supplemented with nystatin. Collected Wasps were subjected to processing, involving serial dilution at a 1:10 dilution (equivalent to a 10-fold dilution), followed by plating on Streptomyces agar for a 7-day incubation period at 37°C. Pure cultures were successfully acquired, streaked onto fresh plates, and then preserved at a temperature of 4°C. Molecular analysis for bacterial genomic DNADNA extraction from bacterial cultures was conducted using Zymo Research’s Quick-DNA™ Fungal/Bacterial Miniprep Kit, involving Proteinase K treatment, centrifugation, and column-based purification. DNA integrity was verified using agarose gel electrophoresis, and DNA yield was quantified with a Qubit 2.0 fluorometer. Molecular identification through PCR utilized primers targeting the 16SrDNA region 27F: 5’AGAGTTTGATCMTGGCTCAG3’) and 1492R: 5’CGGTTACCTTGTTACGACTT3’). Sequencing of PCR products was performed bidirectional using the Nimagen Brilliant Dye™ Terminator Cycle Sequencing Kit V3.1. Sequencing data were analyzed using BioEdit and subjected to BLAST search for taxonomic identification on the NCBI platform. Extraction of bioactive compoundsThe extraction of bioactive metabolites followed a modified method involving acetonitrile and nutrient broth. Briefly. Erlenmeyer flasks were sterilized, inoculated with a sub-cultured isolate, and incubated. After incubation, the medium was decanted, and the solvent was processed through centrifugation and filtration. The resulting solution was transferred to a rotary evaporator for further concentration. Screening of antibacterial activityConcentrated crude acetonitrile extract obtained from the bacteria was screened for their inhibitory activity against 7 known human pathogenic bacterial strains (Escherichia coli from stool, E coli ATCC25922, Listeria monocytogenes ATCC19111, Proteus vulgaris from Abdomen, Staphylococcus aureus ATCC25923, S. aureus from ear and S Pyogenes from sputum.) by cross streak and agar Well Diffusion Assay (Magaldi et al., 2004). Mueller Hinton agar, prepared according to manufacturer’s instructions, was used to seed with an 18-hour culture of each bacterial test strain. Gradient concentrations of 40–100 µl/ml of extracted secondary metabolites were dispensed into labeled holes on the plate using a 6 mm sterilized cork borer. Plates were incubated at 37°C for 18 hours. Acetonitrile served as a negative control, and Ciprofloxacin was used as a positive control. High-performance liquid chromatography (HPLC)HPLC analysis was conducted using Ciprofloxacin as the reference standard. The standard solution was prepared by dissolving 10 mg of ciprofloxacin in 50 ml of acetonitrile with 1% triethylamine, resulting in a concentration of 200 ppm, which was further diluted for analysis. Sample preparation followed a similar protocol, with 20 ml of the sample dissolved in 50 ml of acetonitrile and subsequent dilution. About 5 μl sample was injected onto a C18 column within a chromatographic system equipped with a diode-array detector. Multiple wavelengths were monitored, and separation was achieved through a linear gradient using acetonitrile as the solvent. Concentrations were determined using the equation: Concentration=[area of sample/area of standard] × sample concentration × dilution factor. STRING analysisThe protein-protein interaction network (PPIN) and chemical compounds from the isolates were constructed through the STRING database to predict the interaction and determine the molecular action of these associations and binding affinity (Tiana et al., 2021). ResultsIn this study, 9 isolates were successfully identified at the species level. The identified species included Enterococcus faecalis (OR656467), Pseudomonas fragi (OR150689 and OR150690), Propionibacterium spp (OR150691), Comamonas terrigena (OR656468), Staphylococcus galinarum (OR150693), Levi bacillus brevis (OR150693), Comamonas aquatica (OR656469), and E. faecalis (OR656470). The identity of these isolates ranged from 91.49% to 99.8% with close relatives in the GenBank database (Table 1). Table 1. The organisms identified and their similarity index.
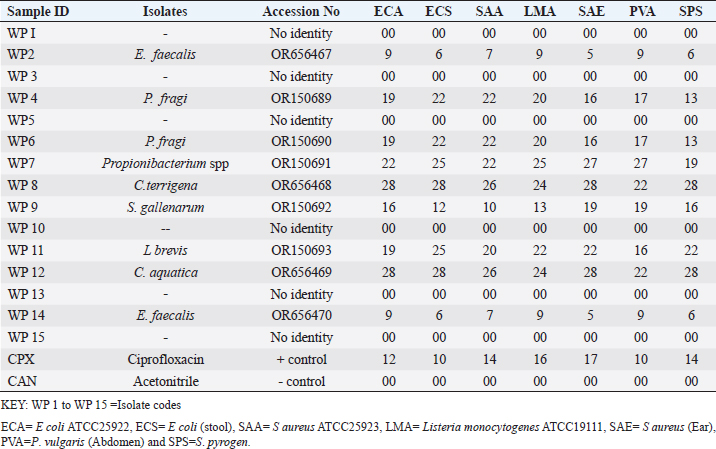
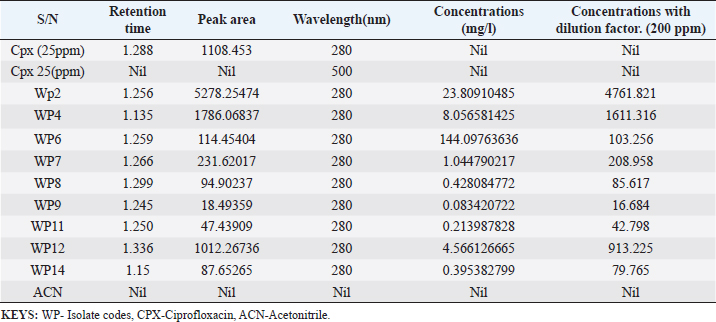
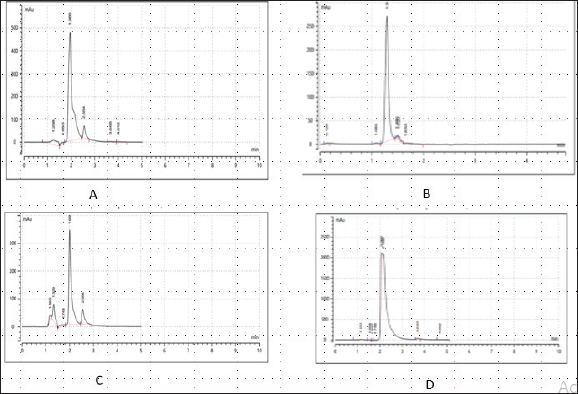
Enterocin produced by E. faecalis (OR656467) exhibited notable inhibition zones, with 9 millimeters against E. coli ATCC25922, L. monocytogenes ATCC19111, and Proteus vulgaris. Similarly, the zone of inhibition ranged from 5 to 7 mm against Staphylococcus aureus obtained from ear, E. coli from stool, and S. aureus ATCC25923. Quinone produced by P. fragi (OR150689) exhibited an inhibition zone of 22 mm against E. coli (from stool) and S. aureus ATCC25923, while a 20 mm zone against Listeria monocytogenes ATCC1911 was measured. Propionicin produced by Propionibacterium (OR150689) exhibited inhibition zones ranging from 25 to 27 mm against four test strains. Metabolites produced by C. terrigena (OR656468) and C. terrigena (OR656469), respectively, demonstrated inhibition zones of 28 mm against S. pyrogen, indicating their consistent antimicrobial activity (Fig. 1). Gallidermin produced by Staphylococcus gallinarum (OR150692) showed inhibition zones of 19 mm against Staphylococcus aureus from obtained from ear and P. vulgaris, as well as 16 mm against S. aureus. Bacteriocins produced by Levi lactobacillus brevis (OR150693) had inhibition zones of 25 mm against E. coli obtained from stool and 22 mm against three test strains, including S. aureus obtained from ear, and L. monocytogenes ATCC19111. A control compound, ciprofloxacin, was also tested, exhibiting varying inhibition zones against the test strains, with the highest being 17 mm against S. aureus obtained from the ear (Table 2). Chromatographic analysisIn this study, the control drug ciprofloxacin (Cpx), analyzed at a wavelength of 280 nm, exhibited a retention time of 1.288 m, a peak area of 1108.453, and unspecified values at 500 nm for retention time, peak area, and concentration. The wasp-associated isolates, specifically WP2, displayed a retention time of 1.256 m, a peak area of 5278.25474, and a concentration of 4761.821 mg/l at 280 nm. WP4 demonstrated a retention time of 1.135 minutes, a peak area of 1786.06837, and a concentration of 1611.316 mg/l at 280 nm. Other isolates, such as WP7, WP8, WP9, WP11, WP12, and WP14, exhibited specific retention times, peak areas, and concentrations at 280 nm (Table 3, Fig. 2). Notably, the retention times of ciprofloxacin (1.288 m) closely aligned with those of the wasp isolates, ranging from 1.135 to 1.336 m. This suggests similarities or close properties between ciprofloxacin and the metabolites produced by the isolates. In contrast, the negative control, acetonitrile, did not show any retention times or peak areas.
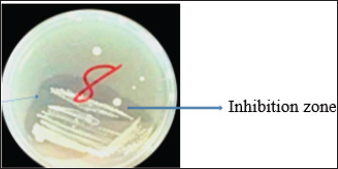
Fig. 1. Inhibition zone produced by Commamonas terrigena against Staphylococcus ATCC25923 by streak method. In-silico analysisIn silico analysis was conducted using the STRING database (Table 4) to examine the protein-protein interactions between the quantified metabolites from wasp-associated bacteria and the test strains. Gallidermin produced by Staphylococcus gallenarium (OR150692) had a positive interaction with S. aureus while Phenazine from P. fragi (OR150689) did not show any interaction with S. aureus and E. coli. Furthermore, bacteriocin from the crude extract of Levilactobaccilus (OR150693) had no positive interaction against S. aureus, E. coli, and L. monocytogenes. In a similar trend, enterocin produced by E. faecalis (OR656467) and E. faecalis (OR656470) did not show matches in the STRING database. The crude extract (Propionicin) obtained from Propionibacterium (OR150691) did not influence the growth of Staphylococcus aureus, E. coli, and L. monocytogenes, respectively. Table 2. Zone of inhibition(mm) of the metabolites against test strains.
Table 3. Quantification and concentration of secondary metabolites obtained from wasp—associated bacteria through HPLC.
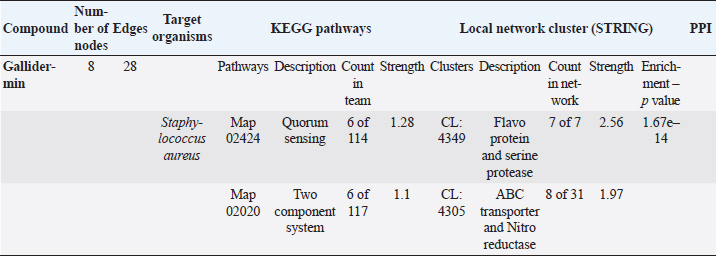
The STRING analysis of gallidermin revealed an interaction network with S. aureus through KEGG pathways and local network clusters. This network comprised 8 nodes and 28 edges associated with gallidermin. The main KEGG pathways identified were Map 02424 for Quorum Sensing (6 nodes out of 114, strength 1.28) and Map 02020 for the two-component system (6 nodes out of 117, strength 1.1). In the local network cluster analysis, a cluster (CL:4349) was associated with flavoprotein and serine protease synthesis. This cluster revealed 7 nodes with a strength of 2.56 and an enrichment p-value of 1.67e–14. Another cluster, CL:4305, was linked to ABC transporter and nitro reductase, with 8 nodes out of 31 and a strength of 1.97. As shown in Table 5 and Figure 3, respectively.
Fig. 2. Chromatograms of secondary metabolites from bacteria associated with wasp. (a) Chromatogram of the control drug (Ciprofloxacin), at 280nm, (b): Chromatogram of the Enterocin extracted from E. faecalis (OR656467) at 280nm, (c): Chromatogram of the phenazine extracted from Pseudomonas fraji (OR150689) at 280 nm and (d): Chromatogram of Gallidermin extracted from Staphylococcus gallinarium (OR150692) at 280 nm. Table 4. Evaluation of clinical potential of secondary metabolites via STRING analysis.
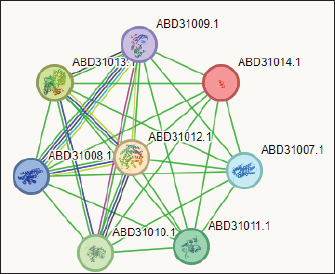
The PPIN comprises 8 nodes representing protein-relevant genes, interconnected by 28 edges that signify interactions between proteins (S. aureus) and bioactive molecules (Gallidermin). The size and color of the nodes convey information about the degree values. Bioactive molecules are depicted by pill-shaped nodes, whereas proteins are represented by spheres. Nodes that share associations are linked by edges, with the length of the lines indicating bond affinities. A shorter edge denotes higher affinity between chemicals and proteins, while a longer one signifies lower affinity. Table 5. Analysis of gallidermin-associated KEGG pathways, and local network cluster.
Fig. 3. The network of interactions of Staphylococcus gallenarum (OR150692) bioactive molecules (Gallidermin, ABD31014.1) with Staphylococcus aureus as predicted by STRING. DiscussionNatural products are now the target for alternative sources of antimicrobial agents especially from microorganisms obtained from soil (Newman and Cragg, 2016). Although, reports have revealed that this could also be isolated from insect-based sources (Oh et al., 2011; Poulsen et al., 2011; Chevrette et al., 2019). In our recent study, we examined a diverse range of bacterial strains isolated from insect-based sources capable of producing secondary metabolites with activity against bacterial pathogens. The results revealed that many insect-based bacteria capable of producing small molecules with activity against bacterial pathogens are eminent and therefore, a source of novel antimicrobial agents. A recent report found that insect-based microbes (Streptomyces) exhibit higher inhibitory activity against fungi, Gram-negative bacteria, and Gram-positive bacteria, compared to both soil and plant-associated strains (Chevrette et al., 2019). The implication is that novel antimicrobials with untapped potentials could be sourced from insect microbes. Poulsen et al. (2011) reported 11 distinct and structurally diverse secondary metabolites from insect-associated Streptomyces with activity against bacteria and fungi. This study although, related in design, but differed in the diversity of organisms and the bioactive compound obtained. There are several reasons for this variation; the research interest of the Authors, the isolation protocol adopted, the composition of the isolation media, probably the differences in geographical location, and other unidentified variables. Furthermore, variations in temperature, humidity, soil composition, and vegetation between different locations can lead to distinct microbial populations and subsequently different bioactive compounds. Likewise, host specificity and species variation among the wasps examined in the studies contribute to the differences observed. Each wasp species harbors a unique microbiome influenced by factors such as diet, nesting behavior, and symbiotic relationships (Receveur et al., 2020). Consequently, the diversity of antibiotic-producing bacteria associated with each species varies, resulting in differences in the bioactive compounds they produce. These findings are significant as they unveil the complexity of microbial communities linked with wasps and highlight the antimicrobial potential of the secondary metabolites against diverse pathogens. Elucidating the underlying mechanisms of metabolite production could further enhance their utility and production in the future. In the present study, the analysis of protein-protein interactions via STRING indicated that gallidermin has a strong positive interaction with S. aureus, suggesting its promise as an effective antibacterial agent. This study corroborates the study conducted by Newstead et al. (2020) who emphasized gallidermin’s ability to inhibit both Staphylococcus epidermidis and S. aureus, including the MRSA strain known for its resistance. Similarly, this work affirmed the conclusions drawn by Bengtsson et al. (2018), in the efficacy of the lantibiotic gallidermin in eradicating S. epidermidis and S. aureus while also mitigating the proinflammatory reactions induced by these bacteria in dermal fibroblasts. Gallidermin’s ability to alleviate inflammatory responses triggered by bacteria in dermal fibroblasts has been reported. Overall, these findings suggest that gallidermin could serve as a potent antimicrobial agent, particularly in combating infections caused by S. aureus, S. epidermidis, and other arrays of pathogenic bacteria. The phenazine secreted by P. fragi showed a favorable interaction with Escherichia coli. However, no other bioactive compounds displayed such interactions with the test strains in our study. This could be largely due to limited information within the STRING database with regard to specific interactions with other microorganisms. In this study, the notable inhibitory effect of phenazine, produced by P. fragi, was prominently observed and is consistent with the report of Lim et al. (2017), who emphasized the substantial antimicrobial capabilities of this compound. Apart from the biotechnological perspective, phenazine compounds have the notable potential to inhibit plant-pathogenic fungi and thus are useful in controlling plant diseases (Bilal et al ., 2017; Lauková et al., 2018). Likewise, enterocins are polypeptides with bacteriostatic activity against important foodborne pathogens, such as S. aureus, Listeria monocytogenes, and Salmonella enterica, and purified enterocin E-760 has been proven to inhibit the growth of 24 species of Gram-negative bacteria and 3 species of Gram-positive bacteria. (Wu et al., 2022). A study reported that the administration of Enterocin M led to the reduction of coliforms and Campylobacter and Clostridium spp (Lauková et al., 2018). The clinical applications of biocin and propionicin (bacterocins) have also been reported in the literature with the tract record of antimicrobial properties against pathogens (Grilli et al., 2009; Atassi and Servin, 2010; Carroll et al., 2010; Campion et al., 2013; Lebel et al., 2013). These findings revealed the inherent treasures that could be harnessed from microbes from diverse sources, particularly the untapped insect-associated sources. ConclusionThe study concluded that wasp-associated bacteria have antimicrobial activity against different pathogenic bacteria. This discovery underscores the potential of exploring additional antimicrobial agents from other insect sources to address the global challenge of drug-resistant pathogens. Given the widespread prevalence of antimicrobial drug resistance, there is an urgent need to expand our investigation into unconventional antimicrobial sources, as they may hold the key to discovering novel treatments and overcoming the current impasse in drug discovery and development. AcknowledgmentThe authors acknowledge all the laboratory scientists of the Department of Microbiology, Babcock University for their technical support. Authors’contributionCOE conceived and designed the study, LSO conducted the study, GNA handled the in-silico analysis, and COE and GNA supervised the study. All authors drafted and proofread the manuscript and agreed to its contents. Data availabilityAll data are provided in the manuscript. Any additional data needed can be provided upon reasonable request from the corresponding author. FundingThis research work was partly funded by the Office of Research Innovation and International Cooperation under the Provost, CPGS Doctoral Thesis Grant. Conflicts of interestThe authors report no financial or any other conflicts of interest in this work. Ethical approvalsEthical clearance was obtained from Babcock University Health Research Ethics Committee (BUHREC-510/22). ReferencesAtassi, F. and Servin, A.L. 2010. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol. Lett. 304, 29–38. Bengtsson, T., Lönn, J., Khalaf, H., and Palm, E. 2018. The lantibiotic gallidermin acts bactericidal against Staphylococcus epidermidis and Staphylococcus aureus and antagonizes the bacteria-induced proinflammatory responses in dermal fibroblasts. Microbiol. Open. 7(6), e00606. Bilal, M., Guo, S., Iqbal, H.M.N., Hu, H., Wang, W. and Zhang, X. 2017. Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: a review. World J Microbiol Biotechnol. 33(10), 191. doi: 10.1007/s11274-017-2356-9 Campion, A., Casey, P.G., Field, D., Cotter, P.D., Hill, C. and Ross R.P. 2013. In vivo activity of Nisin A and Nisin v against Listeria monocytogenes in mice. BMC Microbiol. 13, 23. Carroll, J., Draper, L.A., O’Connor, P.M., Coffey, A., Hill, C., Ross, R.P., Cotter, P.D. and O’Mahony, J. 2010. Comparison of the activities of the lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int. J. Antimicrob. Agents. 36, 132–136. Chevrette, M.G., Carlson, C.M., Ortega, H.E, Thomas, C., Ananiev, G.E., Barns, K.J., Book, A.J., Cagnazzo, J., Carlos, C., Flanigan, W., Grubbs, K.J., Horn, H.A., Hoffmann, F.M., Klassen, J.L., Knack, J.J., Lewin, G.R., McDonald, B.R., Muller, L., Melo, W.G.P., Pinto-Tomás, A.A,. Schmitz, A., Wendt-Pienkowski, E., Wildman, S., Zhao, M., Zhang, F., Bugni, T.S., Andes, D.R., Pupo, M.T. and Currie, C.R. 2019. The antimicrobial potential of Streptomyces from insect microbiomes. NA. Commun. 10, 516 | Available via https://doi.org/10.1038/s41467-019-08438-0 Grilli, E., Messina, M., Catelli, E., Morlacchini, M. and Piva, A. 2009. Pediocin improves growth performance of broilers challenged with Clostridium perfringens. Poult. Sci. 88, 2152–2158. Imran, M.D., Kirti, D.R. and Naik, M.M. 2018. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Environ. Pollut 233, 132. Lauková, A., Styková, E., Kubašová, I., Gancarčíková, S., Plachá, I., Mudroňová, D., Kandričáková, A., Miltko, R., Belzecki, G. and Valocký, I. 2018. Enterocin M and its beneficial effects in horses-a pilot experiment. Probiotics Antimicrob. Proteins. 10, 420–426. Laxminarayan, R. 2022. The overlooked pandemic of antimicrobial resistance. Lancet. 399, 606–607. Lebel, G., Piché, F., Frenette, M., Gottschalk, M. and Grenier, D. 2013. Antimicrobial activity of nisin against the swine pathogen Streptococcus suis and its synergistic interaction with antibiotics. Peptides. 50, 19–23. Lim, D.J., Yang, S.Y., Noh, M.Y., Lee, C.W., Kim, J.C. and Kim, I.S. 2017. Identification of lipopeptide xantholysins from Pseudomonas sp. DJ15 and their insecticidal activity against Myzus persicae. Entomol. Res. 47, 337–343. Magaldi, S., Mata-Essayag, S., Hartung de Capriles, C., Perez, C., Colella, MT., Olaizola, C. and Ontiveros, Y. 2004. Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 8(1), 39–45. Murray, C.J., Ikuta, K.S, Sharara, F., Swetschinski, L., Aguilar, GR., Gray, A., Han, C., Bisignano, C., Rao, P., Wool, E. and Johnson SC. 2022. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 399, 629–655. Newman, D.J. and Cragg, G.M. 2016. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. Newstead, L.L., Varjonen, K., Nuttall, T. and Paterson, G.K. 2020. Staphylococcal-produced bacteriocins and antimicrobial peptides. Antibiotics. 9(2), 40. Nwobodo, D.C, Ugwu, M.C., Anie, C.O., Al-Ouqaili, M.T.S., Ikem, J.C., Chigozie, U.V. and Saki, M. 2022. Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 36(9), e24655. Oh, D.C., Poulsen, M., Currie, C.R. and Clardy, J. 2011. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org Lett. 13(4), 752–755. Oliver, S.P., Murinda, S.E. and Jayarao, B.M. 2011. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: a comprehensive review. Foodborne Pathog. Dis. 8(3), 337–355. Poulsen, M., Oh, D.C., Clardy, J. and Currie, C.R. 2011. Chemical analyses of wasp associated Streptomyces bacteria reveal a prolific potential for natural products discovery. Environ. Microbiol. Rep. 2, 534–540. Receveur, J.P., Fenoglio, S. and Benbow, M.E. 2020. Insect-associated bacterial communities in an alpine stream. Hydrobiologia. 847, 331–344. Tiana, C., Liua., X, Changa Y., Wanga, R., Lva, T., Cuia, C. and Liua M. 2021. Investigation of the anti-inflammatory and antioxidant activities of luteolin, kaempferol, apigenin and quercetin. S. Afr. J. Bot. 137, 257–264. Wu, Y., Pang. X., Wu, Y., Liu, X. and Zhang X. 2022. Enterocins: classification, synthesis, antibacterial mechanisms and food applications. Molecules. 27, 2258. Available via https://doi.org/10.3390/ molecules27072258 | ||
| How to Cite this Article |
| Pubmed Style Owolabi LS, Ezeamagu CO, Anyasor GN. Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. J Microbiol Infect Dis. 2024; 14(2): 80-87. doi:10.5455/JMID.2024.v14.i2.6 Web Style Owolabi LS, Ezeamagu CO, Anyasor GN. Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. https://www.jmidonline.org/?mno=196205 [Access: January 25, 2026]. doi:10.5455/JMID.2024.v14.i2.6 AMA (American Medical Association) Style Owolabi LS, Ezeamagu CO, Anyasor GN. Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. J Microbiol Infect Dis. 2024; 14(2): 80-87. doi:10.5455/JMID.2024.v14.i2.6 Vancouver/ICMJE Style Owolabi LS, Ezeamagu CO, Anyasor GN. Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. J Microbiol Infect Dis. (2024), [cited January 25, 2026]; 14(2): 80-87. doi:10.5455/JMID.2024.v14.i2.6 Harvard Style Owolabi, L. S., Ezeamagu, . C. O. & Anyasor, . G. N. (2024) Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. J Microbiol Infect Dis, 14 (2), 80-87. doi:10.5455/JMID.2024.v14.i2.6 Turabian Style Owolabi, Lawrence Shola, Cajethan Onyebuchi Ezeamagu, and Godswill Nduka Anyasor. 2024. Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. Journal of Microbiology and Infectious Diseases, 14 (2), 80-87. doi:10.5455/JMID.2024.v14.i2.6 Chicago Style Owolabi, Lawrence Shola, Cajethan Onyebuchi Ezeamagu, and Godswill Nduka Anyasor. "Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria." Journal of Microbiology and Infectious Diseases 14 (2024), 80-87. doi:10.5455/JMID.2024.v14.i2.6 MLA (The Modern Language Association) Style Owolabi, Lawrence Shola, Cajethan Onyebuchi Ezeamagu, and Godswill Nduka Anyasor. "Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria." Journal of Microbiology and Infectious Diseases 14.2 (2024), 80-87. Print. doi:10.5455/JMID.2024.v14.i2.6 APA (American Psychological Association) Style Owolabi, L. S., Ezeamagu, . C. O. & Anyasor, . G. N. (2024) Antibacterial properties of secondary metabolites produced from wasp-associated bacteria in Isara, Remo-North, Ogun State, Nigeria. Journal of Microbiology and Infectious Diseases, 14 (2), 80-87. doi:10.5455/JMID.2024.v14.i2.6 |